class: bottom, left, inverse, title-slide # Surviving Data Analysis ## An Evaluator’s Gentle Introduction to R ### Petr Bouchal ### 8 June 2021 --- class: left, middle, inverse # Welcome! --- class: left, large, inverse # Who I am social scientist by background worked in public service, think tanks, consulting no technical education beyond an economics degree now focus on quality of institutions, data analysis, public finances --- class: left, middle, inverse # Why we are here today --- class: left, top, large 1\. Code + data = more manageable projects -- 2\. Get a sense of when and why to use R -- 3\. Find our way around R and RStudio -- 4\. Hands-on: common data analysis tasks in R -- 5\. Use R to make data analysis projects manageable -- 6\. Know where to go to learn more --- class: left, large # What we are not doing making you an expert in each of the tools learning statistics talking about big data, AI and other buzzwords --- class: left, middle, inverse, large # The plan Motivation: introducing the case study Manageable data analysis projects A gentle intro to R Dissecting the case study: tools & practices After we part ??? The case study will provide an overview of the core components of the R toolkit: - writing code - loading and analysing data - visualising data - blending code and text into automated outputs - organising the project We don't start with teaching you the syntax and usage of R: - so we don't get bogged down in detail - so you understand useful bits in use in the case study (tidyverse) By the end of today, you won't necessarily understand every line or be able to create it, but you will understand what are the different tools that are needed to build a project like this and where to learn to use them in more depth. --- class: left, middle, inverse # But first: light technical setup --- class: center, middle .large[ [petrbouchal.xyz/czecheval2021](https://petrbouchal.xyz/czecheval2021) ] --- class: left, middle, medium - Go to [rstudio.cloud](https://rstudio.cloud) - Log in the way you signed up (email, Google account, Github account) - "New project > New Project from Git Repository"  - enter https://github.com/petrbouchal/czecheval2021 - Run `renv::restore()` in Console ??? https://rstudio.cloud https://github.com/petrbouchal/czecheval2021 renv::restore() --- class: left, middle, inverse # Motivation --- class: left, middle, large open `report.Rmd` click blue "Knit" button on the toolbar > "Knit to HTML" ??? Output - text and code - output: all in one - numbers are update automatically Directory: - Rmd source - input data - output data Together, these allow you to run what I call manageable projects. --- class: middle, inverse, large # *Manageable data analysis projects* # or # what we want from data analysis --- class: large, middle Only do valuable work and do it efficiently so that you don't go mad, nor your future self or your colleagues and so that you can be sure the results are right. --- class: medium .pull-left[ ## What a good project should be like - automated - transparent - easily tweaked - easily repeated - low friction - (easy to collaborate on in parallel or in sequence) - (clear history of changes) ] -- .pull-right[ ## What it means in practice - code + data - self-contained - documented - well organised (code, files, workflow) - code + text - (version control) ] --- class: medium, middle, center ad-hoc calculation ... **repeatable/automated report** reproducible report ... production data system ??? -There is a spectrum -There are nuances - replication, repetition, reproducible, automated In different contexts, you will want different standards, but having at least basic reproducibility (i.e. the code will work if someone else runs in order) is good even for the most basic mini projects. --- class: large # Why use R for this .pull-left.center.middle[ Open source Flexibility *Community* ] .pull-right.center.middle[ Data Visualisation and reporting *RStudio* ] ??? Versus Python? By statistitians, for data Community = inclusive, ppl. w/varied backgrounds, non-programmers Versatile: I show you tabular and spatial data, but you can work with anything. Very good for any kind of modeling. Used in critical and high-profile applications (science, newsrooms, industry, government). --- class: middle, inverse, large # Dissecting the case study --- class: large 1\. Data + code + text = data analysis 2\. Running code, basic operations 3\. Working with tabular data 4\. Data visualisation 5\. Making projects manageable --- class: large **1\. Data + code + text = data analysis** 2\. Running code, basic operations 3\. Working with tabular data 4\. Data visualisation 5\. Making projects manageable --- class: middle, large # A tour of your screen ## RStudio .pull-left[ 1. Scripts 1. Console ] .pull-right[ 1. Environment + History 1. Files + Plots + Packages + Help + Viewer ] ??? We will now take a look at simpler examples of the two core components: - R code - markdown --- class: large 1\. Data + code + text = data analysis **2\. Running code, basic operations** 3\. Working with tabular data 4\. Data visualisation 5\. Making projects manageable --- class: center, middle, large open `example.md` --- class: center, middle, large open `example.Rmd` --- class: large # Core concepts: Markdown + Rmarkdown .pull-left[ markdown - plain text - YAML header - source versus output - Visual Editor ] .pull-right[ Rmarkdown - code chunk - chunk options - inline code - output formats - knitting ] --- class: center, middle, large open `example.R` ??? Autocomplete => how to name variables --- class: large # Aside: seeking help - `?function` / `??anything` - click a function in a script, then F1 - "Google the error message" - Stack Overflow - RStudio Community - package documentation: vignettes and online - [RStudio Cheatsheets](https://rstudio.com/resources/cheatsheets/) --- class: large, middle, center # Core concepts: R .pull-left[ object function variable data.frame ] .pull-right[ package script environment ] --- class: medium # Summary: what is what in R "Only the data and code are real" .pull-left[ ## Persistent/real - code & text (R or Rmarkdown document) - input data - (other input files, like images) ] .pull-right[ ## Transient - outputs: documents, chart files, output data files - objects in the R environment ] see https://socviz.co/gettingstarted.html#things-to-know-about-r --- ### Input data is not altered by code Data are read from a file into the R environmet. We use code to operate on the data in the environment, not on the data file. The result can be written into a new file as needed. ### Everything I do to the data must be recorded in code No manual editing of the input data files. No manual editing of output files (only at the end if needed) The code file should run from start to finish without errors. ### What is in the Environment does not persist Once R is restarted, it disappears (restart often!) If you want to keep it, you must have a way of reproducing it with code. --- class: center, middle, large Following these principles will make your data work more reproducible. These also apply to data analysis in other languages (Python, Julia aj.) --- class: large 1\. Data + code + text = data analysis 2\. Running code, basic operations **3\. Working with tabular data** 4\. Data visualisation 5\. Making projects manageable --- class: large # Tidyverse: what it is - set of R packages with consistent usage principles - built for tabular data (rows and columns) - grammar analogy: a set of verbs for each type of tasks - follows logic of data analysis - can be thought of as extension or dialect of R --- class: left, top # Tidyverse: the basic logic .center.middle[ 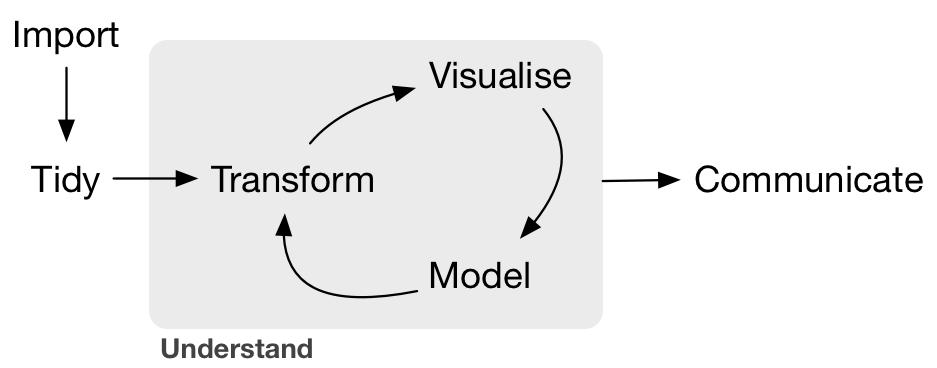 ] --- class: large # A package for each set of tasks .center[ <img src="tidyverse-logos.png" width="60%" /> ] --- class: large # A package for each set of tasks - `readr` load data from text files (CSV, TSV) - `readxl`: import data from Excel - `dplyr`: basic data manipulation - `tidyr` data cleaning and reshaping - `stringr`: work with texts ("characters"/"strings") - `lubridate` work with dates and times - `ggplot2`: data visualisation --- class: large, center, middle [dplyr.tidyverse.org](https://dplyr.tidyverse.org) [readr.tidyverse.org](https://dplyr.tidyverse.org) ... Reference | Articles --- ## Further: - `forcats` for working with factors (categorical data) - `httr` a `rvest` for accessing and scraping web data - `tibble` for handling data frames - `glue` for concatenating strings and data in a neat way - `purrr` for looping through operations across data ## Even more: - `haven` for loading SPSS, Stata and other data - `DBI`, `dbplyr` and friends: interface to databases - `writexl` for exporting data into excel - `sf` for working with spatial (geographical) data --- class: medium *Verbs* in the tidyverse (`dplyr` and `tidyr` packages) - `filter`: select rows that fit a rule - `mutate`: calculate new columns - `summarise`: summarise using a summary function (sum, mean, ...) - `group_by`: run operations for each group - `select`: select columns (as in SQL) - `arrange`: ordering rows based on one or multiple columns - `join_*:` join multiple datasets based on a key column - `pivot_*:` reshaping ("long" <=> "wide" form) --- class: large Other useful functions - `count` - `starts_with` / `ends_with` / `matches` - `distinct` - `n_distinct` - `rename` - `recode` - `separate` / `unite` - `bind_rows` --- class: center, middle, large # Tidy data manipulation --- count: false .panel1-tidyverse-example-auto[ ```r *vd ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 61,898 x 8 date vaccine nuts3 nuts3_name vekova_skupina prvnich_davek <date> <chr> <chr> <chr> <chr> <dbl> 1 2020-12-27 Comirnaty CZ010 Hlavní město Praha 18-24 49 2 2020-12-27 Comirnaty CZ010 Hlavní město Praha 25-29 110 3 2020-12-27 Comirnaty CZ010 Hlavní město Praha 30-34 102 4 2020-12-27 Comirnaty CZ010 Hlavní město Praha 35-39 111 5 2020-12-27 Comirnaty CZ010 Hlavní město Praha 40-44 169 6 2020-12-27 Comirnaty CZ010 Hlavní město Praha 45-49 156 7 2020-12-27 Comirnaty CZ010 Hlavní město Praha 50-54 128 8 2020-12-27 Comirnaty CZ010 Hlavní město Praha 55-59 96 9 2020-12-27 Comirnaty CZ010 Hlavní město Praha 60-64 85 10 2020-12-27 Comirnaty CZ010 Hlavní město Praha 65-69 79 11 2020-12-27 Comirnaty CZ010 Hlavní město Praha 70-74 48 12 2020-12-27 Comirnaty CZ010 Hlavní město Praha 75-79 19 13 2020-12-27 Comirnaty CZ010 Hlavní město Praha 80+ 24 14 2020-12-27 Comirnaty CZ064 Jihomoravský kraj 25-29 3 15 2020-12-27 Comirnaty CZ064 Jihomoravský kraj 30-34 7 16 2020-12-27 Comirnaty CZ064 Jihomoravský kraj 35-39 8 17 2020-12-27 Comirnaty CZ064 Jihomoravský kraj 40-44 6 18 2020-12-27 Comirnaty CZ064 Jihomoravský kraj 45-49 10 19 2020-12-27 Comirnaty CZ064 Jihomoravský kraj 50-54 14 20 2020-12-27 Comirnaty CZ064 Jihomoravský kraj 55-59 11 # … with 61,878 more rows, and 2 more variables: druhych_davek <dbl>, # doses_total <dbl> ``` ] --- count: false .panel1-tidyverse-example-auto[ ```r vd %>% * select(date, vaccine, nuts3, doses_total) ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 61,898 x 4 date vaccine nuts3 doses_total <date> <chr> <chr> <dbl> 1 2020-12-27 Comirnaty CZ010 49 2 2020-12-27 Comirnaty CZ010 110 3 2020-12-27 Comirnaty CZ010 102 4 2020-12-27 Comirnaty CZ010 111 5 2020-12-27 Comirnaty CZ010 169 6 2020-12-27 Comirnaty CZ010 156 7 2020-12-27 Comirnaty CZ010 128 8 2020-12-27 Comirnaty CZ010 96 9 2020-12-27 Comirnaty CZ010 85 10 2020-12-27 Comirnaty CZ010 79 11 2020-12-27 Comirnaty CZ010 48 12 2020-12-27 Comirnaty CZ010 19 13 2020-12-27 Comirnaty CZ010 24 14 2020-12-27 Comirnaty CZ064 3 15 2020-12-27 Comirnaty CZ064 7 16 2020-12-27 Comirnaty CZ064 8 17 2020-12-27 Comirnaty CZ064 6 18 2020-12-27 Comirnaty CZ064 10 19 2020-12-27 Comirnaty CZ064 14 20 2020-12-27 Comirnaty CZ064 11 # … with 61,878 more rows ``` ] --- count: false .panel1-tidyverse-example-auto[ ```r vd %>% select(date, vaccine, nuts3, doses_total) %>% * mutate(month = lubridate::floor_date(date, "month")) ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 61,898 x 5 date vaccine nuts3 doses_total month <date> <chr> <chr> <dbl> <date> 1 2020-12-27 Comirnaty CZ010 49 2020-12-01 2 2020-12-27 Comirnaty CZ010 110 2020-12-01 3 2020-12-27 Comirnaty CZ010 102 2020-12-01 4 2020-12-27 Comirnaty CZ010 111 2020-12-01 5 2020-12-27 Comirnaty CZ010 169 2020-12-01 6 2020-12-27 Comirnaty CZ010 156 2020-12-01 7 2020-12-27 Comirnaty CZ010 128 2020-12-01 8 2020-12-27 Comirnaty CZ010 96 2020-12-01 9 2020-12-27 Comirnaty CZ010 85 2020-12-01 10 2020-12-27 Comirnaty CZ010 79 2020-12-01 11 2020-12-27 Comirnaty CZ010 48 2020-12-01 12 2020-12-27 Comirnaty CZ010 19 2020-12-01 13 2020-12-27 Comirnaty CZ010 24 2020-12-01 14 2020-12-27 Comirnaty CZ064 3 2020-12-01 15 2020-12-27 Comirnaty CZ064 7 2020-12-01 16 2020-12-27 Comirnaty CZ064 8 2020-12-01 17 2020-12-27 Comirnaty CZ064 6 2020-12-01 18 2020-12-27 Comirnaty CZ064 10 2020-12-01 19 2020-12-27 Comirnaty CZ064 14 2020-12-01 20 2020-12-27 Comirnaty CZ064 11 2020-12-01 # … with 61,878 more rows ``` ] --- count: false .panel1-tidyverse-example-auto[ ```r vd %>% select(date, vaccine, nuts3, doses_total) %>% mutate(month = lubridate::floor_date(date, "month")) %>% * group_by(month, vaccine, nuts3) ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 61,898 x 5 # Groups: month, vaccine, nuts3 [280] date vaccine nuts3 doses_total month <date> <chr> <chr> <dbl> <date> 1 2020-12-27 Comirnaty CZ010 49 2020-12-01 2 2020-12-27 Comirnaty CZ010 110 2020-12-01 3 2020-12-27 Comirnaty CZ010 102 2020-12-01 4 2020-12-27 Comirnaty CZ010 111 2020-12-01 5 2020-12-27 Comirnaty CZ010 169 2020-12-01 6 2020-12-27 Comirnaty CZ010 156 2020-12-01 7 2020-12-27 Comirnaty CZ010 128 2020-12-01 8 2020-12-27 Comirnaty CZ010 96 2020-12-01 9 2020-12-27 Comirnaty CZ010 85 2020-12-01 10 2020-12-27 Comirnaty CZ010 79 2020-12-01 11 2020-12-27 Comirnaty CZ010 48 2020-12-01 12 2020-12-27 Comirnaty CZ010 19 2020-12-01 13 2020-12-27 Comirnaty CZ010 24 2020-12-01 14 2020-12-27 Comirnaty CZ064 3 2020-12-01 15 2020-12-27 Comirnaty CZ064 7 2020-12-01 16 2020-12-27 Comirnaty CZ064 8 2020-12-01 17 2020-12-27 Comirnaty CZ064 6 2020-12-01 18 2020-12-27 Comirnaty CZ064 10 2020-12-01 19 2020-12-27 Comirnaty CZ064 14 2020-12-01 20 2020-12-27 Comirnaty CZ064 11 2020-12-01 # … with 61,878 more rows ``` ] --- count: false .panel1-tidyverse-example-auto[ ```r vd %>% select(date, vaccine, nuts3, doses_total) %>% mutate(month = lubridate::floor_date(date, "month")) %>% group_by(month, vaccine, nuts3) %>% * summarise(doses_total_monthly = sum(doses_total)) ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 280 x 4 # Groups: month, vaccine [21] month vaccine nuts3 doses_total_monthly <date> <chr> <chr> <dbl> 1 2020-12-01 Comirnaty CZ010 5522 2 2020-12-01 Comirnaty CZ020 19 3 2020-12-01 Comirnaty CZ032 15 4 2020-12-01 Comirnaty CZ041 1 5 2020-12-01 Comirnaty CZ042 147 6 2020-12-01 Comirnaty CZ053 11 7 2020-12-01 Comirnaty CZ063 1 8 2020-12-01 Comirnaty CZ064 5026 9 2020-12-01 Comirnaty CZ071 211 10 2020-12-01 Comirnaty CZ080 816 11 2021-01-01 Comirnaty CZ010 67937 12 2021-01-01 Comirnaty CZ020 21110 13 2021-01-01 Comirnaty CZ031 18702 14 2021-01-01 Comirnaty CZ032 13483 15 2021-01-01 Comirnaty CZ041 5431 16 2021-01-01 Comirnaty CZ042 11138 17 2021-01-01 Comirnaty CZ051 7964 18 2021-01-01 Comirnaty CZ052 14203 19 2021-01-01 Comirnaty CZ053 8470 20 2021-01-01 Comirnaty CZ063 9903 # … with 260 more rows ``` ] --- count: false .panel1-tidyverse-example-auto[ ```r vd %>% select(date, vaccine, nuts3, doses_total) %>% mutate(month = lubridate::floor_date(date, "month")) %>% group_by(month, vaccine, nuts3) %>% summarise(doses_total_monthly = sum(doses_total)) %>% * ungroup() ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 280 x 4 month vaccine nuts3 doses_total_monthly <date> <chr> <chr> <dbl> 1 2020-12-01 Comirnaty CZ010 5522 2 2020-12-01 Comirnaty CZ020 19 3 2020-12-01 Comirnaty CZ032 15 4 2020-12-01 Comirnaty CZ041 1 5 2020-12-01 Comirnaty CZ042 147 6 2020-12-01 Comirnaty CZ053 11 7 2020-12-01 Comirnaty CZ063 1 8 2020-12-01 Comirnaty CZ064 5026 9 2020-12-01 Comirnaty CZ071 211 10 2020-12-01 Comirnaty CZ080 816 11 2021-01-01 Comirnaty CZ010 67937 12 2021-01-01 Comirnaty CZ020 21110 13 2021-01-01 Comirnaty CZ031 18702 14 2021-01-01 Comirnaty CZ032 13483 15 2021-01-01 Comirnaty CZ041 5431 16 2021-01-01 Comirnaty CZ042 11138 17 2021-01-01 Comirnaty CZ051 7964 18 2021-01-01 Comirnaty CZ052 14203 19 2021-01-01 Comirnaty CZ053 8470 20 2021-01-01 Comirnaty CZ063 9903 # … with 260 more rows ``` ] --- count: false .panel1-tidyverse-example-auto[ ```r vd %>% select(date, vaccine, nuts3, doses_total) %>% mutate(month = lubridate::floor_date(date, "month")) %>% group_by(month, vaccine, nuts3) %>% summarise(doses_total_monthly = sum(doses_total)) %>% ungroup() %>% * group_by(vaccine, nuts3) ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 280 x 4 # Groups: vaccine, nuts3 [56] month vaccine nuts3 doses_total_monthly <date> <chr> <chr> <dbl> 1 2020-12-01 Comirnaty CZ010 5522 2 2020-12-01 Comirnaty CZ020 19 3 2020-12-01 Comirnaty CZ032 15 4 2020-12-01 Comirnaty CZ041 1 5 2020-12-01 Comirnaty CZ042 147 6 2020-12-01 Comirnaty CZ053 11 7 2020-12-01 Comirnaty CZ063 1 8 2020-12-01 Comirnaty CZ064 5026 9 2020-12-01 Comirnaty CZ071 211 10 2020-12-01 Comirnaty CZ080 816 11 2021-01-01 Comirnaty CZ010 67937 12 2021-01-01 Comirnaty CZ020 21110 13 2021-01-01 Comirnaty CZ031 18702 14 2021-01-01 Comirnaty CZ032 13483 15 2021-01-01 Comirnaty CZ041 5431 16 2021-01-01 Comirnaty CZ042 11138 17 2021-01-01 Comirnaty CZ051 7964 18 2021-01-01 Comirnaty CZ052 14203 19 2021-01-01 Comirnaty CZ053 8470 20 2021-01-01 Comirnaty CZ063 9903 # … with 260 more rows ``` ] --- count: false .panel1-tidyverse-example-auto[ ```r vd %>% select(date, vaccine, nuts3, doses_total) %>% mutate(month = lubridate::floor_date(date, "month")) %>% group_by(month, vaccine, nuts3) %>% summarise(doses_total_monthly = sum(doses_total)) %>% ungroup() %>% group_by(vaccine, nuts3) %>% * mutate(doses_total_monthly_cum = cumsum(doses_total_monthly)) ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 280 x 5 # Groups: vaccine, nuts3 [56] month vaccine nuts3 doses_total_monthly doses_total_monthly_cum <date> <chr> <chr> <dbl> <dbl> 1 2020-12-01 Comirnaty CZ010 5522 5522 2 2020-12-01 Comirnaty CZ020 19 19 3 2020-12-01 Comirnaty CZ032 15 15 4 2020-12-01 Comirnaty CZ041 1 1 5 2020-12-01 Comirnaty CZ042 147 147 6 2020-12-01 Comirnaty CZ053 11 11 7 2020-12-01 Comirnaty CZ063 1 1 8 2020-12-01 Comirnaty CZ064 5026 5026 9 2020-12-01 Comirnaty CZ071 211 211 10 2020-12-01 Comirnaty CZ080 816 816 11 2021-01-01 Comirnaty CZ010 67937 73459 12 2021-01-01 Comirnaty CZ020 21110 21129 13 2021-01-01 Comirnaty CZ031 18702 18702 14 2021-01-01 Comirnaty CZ032 13483 13498 15 2021-01-01 Comirnaty CZ041 5431 5432 16 2021-01-01 Comirnaty CZ042 11138 11285 17 2021-01-01 Comirnaty CZ051 7964 7964 18 2021-01-01 Comirnaty CZ052 14203 14203 19 2021-01-01 Comirnaty CZ053 8470 8481 20 2021-01-01 Comirnaty CZ063 9903 9904 # … with 260 more rows ``` ] --- count: false .panel1-tidyverse-example-auto[ ```r vd %>% select(date, vaccine, nuts3, doses_total) %>% mutate(month = lubridate::floor_date(date, "month")) %>% group_by(month, vaccine, nuts3) %>% summarise(doses_total_monthly = sum(doses_total)) %>% ungroup() %>% group_by(vaccine, nuts3) %>% mutate(doses_total_monthly_cum = cumsum(doses_total_monthly)) %>% * arrange(nuts3, vaccine, month) ``` ] .panel2-tidyverse-example-auto[ ``` # A tibble: 280 x 5 # Groups: vaccine, nuts3 [56] month vaccine nuts3 doses_total_monthly doses_total_monthly… <date> <chr> <chr> <dbl> <dbl> 1 2020-12-01 Comirnaty CZ010 5522 5522 2 2021-01-01 Comirnaty CZ010 67937 73459 3 2021-02-01 Comirnaty CZ010 75930 149389 4 2021-03-01 Comirnaty CZ010 124294 273683 5 2021-04-01 Comirnaty CZ010 158353 432036 6 2021-05-01 Comirnaty CZ010 299988 732024 7 2021-06-01 Comirnaty CZ010 57208 789232 8 2021-04-01 COVID-19 Vaccine J… CZ010 660 660 9 2021-05-01 COVID-19 Vaccine J… CZ010 4256 4916 10 2021-06-01 COVID-19 Vaccine J… CZ010 1477 6393 11 2021-01-01 COVID-19 Vaccine M… CZ010 1 1 12 2021-02-01 COVID-19 Vaccine M… CZ010 2 3 13 2021-03-01 COVID-19 Vaccine M… CZ010 17060 17063 14 2021-04-01 COVID-19 Vaccine M… CZ010 23557 40620 15 2021-05-01 COVID-19 Vaccine M… CZ010 25304 65924 16 2021-06-01 COVID-19 Vaccine M… CZ010 5297 71221 17 2021-02-01 VAXZEVRIA CZ010 1280 1280 18 2021-03-01 VAXZEVRIA CZ010 20688 21968 19 2021-04-01 VAXZEVRIA CZ010 16838 38806 20 2021-05-01 VAXZEVRIA CZ010 18308 57114 # … with 260 more rows ``` ] <style> .panel1-tidyverse-example-auto { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-tidyverse-example-auto { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-tidyverse-example-auto { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- class: large 1\. Data + code + text = data analysis 2\. Running code, basic operations 3\. Working with tabular data **4\. Data visualisation** 5\. Making projects manageable --- class: center, middle, large # ggplot2: The Grammar of Graphics --- count: false .panel1-ggplot2-example-non_seq[ ```r ggplot(data = vd_reg_month, ) + labs(title = "Total doses by region and vaccine type", subtitle = "Cumulative monthly totals") + theme( ) ``` ] .panel2-ggplot2-example-non_seq[ <img src="index_files/figure-html/ggplot2-example_non_seq_01_output-1.png" width="504" /> ] --- count: false .panel1-ggplot2-example-non_seq[ ```r ggplot(data = vd_reg_month, * mapping = aes(x = month, y = doses_total_cum) ) + labs(title = "Total doses by region and vaccine type", subtitle = "Cumulative monthly totals") + theme( ) ``` ] .panel2-ggplot2-example-non_seq[ <img src="index_files/figure-html/ggplot2-example_non_seq_02_output-1.png" width="504" /> ] --- count: false .panel1-ggplot2-example-non_seq[ ```r ggplot(data = vd_reg_month, mapping = aes(x = month, y = doses_total_cum) ) + * geom_col( * ) + labs(title = "Total doses by region and vaccine type", subtitle = "Cumulative monthly totals") + theme( ) ``` ] .panel2-ggplot2-example-non_seq[ <img src="index_files/figure-html/ggplot2-example_non_seq_03_output-1.png" width="504" /> ] --- count: false .panel1-ggplot2-example-non_seq[ ```r ggplot(data = vd_reg_month, mapping = aes(x = month, y = doses_total_cum) ) + geom_col( * aes(fill = vaccine) ) + labs(title = "Total doses by region and vaccine type", subtitle = "Cumulative monthly totals") + theme( ) ``` ] .panel2-ggplot2-example-non_seq[ <img src="index_files/figure-html/ggplot2-example_non_seq_04_output-1.png" width="504" /> ] --- count: false .panel1-ggplot2-example-non_seq[ ```r ggplot(data = vd_reg_month, mapping = aes(x = month, y = doses_total_cum) ) + geom_col( aes(fill = vaccine) ) + * facet_wrap(vars(nuts3_name)) + labs(title = "Total doses by region and vaccine type", subtitle = "Cumulative monthly totals") + theme( ) ``` ] .panel2-ggplot2-example-non_seq[ <img src="index_files/figure-html/ggplot2-example_non_seq_05_output-1.png" width="504" /> ] --- count: false .panel1-ggplot2-example-non_seq[ ```r ggplot(data = vd_reg_month, mapping = aes(x = month, y = doses_total_cum) ) + geom_col( aes(fill = vaccine) ) + facet_wrap(vars(nuts3_name)) + * scale_fill_brewer(palette = "Set1") + labs(title = "Total doses by region and vaccine type", subtitle = "Cumulative monthly totals") + theme( ) ``` ] .panel2-ggplot2-example-non_seq[ <img src="index_files/figure-html/ggplot2-example_non_seq_06_output-1.png" width="504" /> ] --- count: false .panel1-ggplot2-example-non_seq[ ```r ggplot(data = vd_reg_month, mapping = aes(x = month, y = doses_total_cum) ) + geom_col( aes(fill = vaccine) ) + facet_wrap(vars(nuts3_name)) + scale_fill_brewer(palette = "Set1") + labs(title = "Total doses by region and vaccine type", subtitle = "Cumulative monthly totals") + theme( * legend.position = "bottom" ) ``` ] .panel2-ggplot2-example-non_seq[ <img src="index_files/figure-html/ggplot2-example_non_seq_07_output-1.png" width="504" /> ] <style> .panel1-ggplot2-example-non_seq { color: black; width: 38.6060606060606%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel2-ggplot2-example-non_seq { color: black; width: 59.3939393939394%; hight: 32%; float: left; padding-left: 1%; font-size: 80% } .panel3-ggplot2-example-non_seq { color: black; width: NA%; hight: 33%; float: left; padding-left: 1%; font-size: 80% } </style> --- Assign variables/columns to aestherics in `mapping = aes(...)`; asign fixed values (like `colour = "red"`) outside `aes()`. .pull-left[ ```r ggplot(data = vd_reg_month, mapping = aes(x = month, y = doses_total)) + * geom_col(aes(fill = vaccine)) ``` <img src="index_files/figure-html/unnamed-chunk-5-1.png" width="504" /> ] .pull-right[ ```r ggplot(data = vd_reg_month, mapping = aes(x = month, y = doses_total)) + * geom_col(fill = "red") ``` <img src="index_files/figure-html/unnamed-chunk-6-1.png" width="504" /> ] --- class: large 1\. Data + code + text = data analysis 2\. Running code, basic operations 3\. Working with tabular data 4\. Data visualisation **5\. Making projects manageable** --- class: medium Think through the purpose: automation, adaptation, repeatability, reproducibility .pull-left[ ## Good practice and discipline - [naming things](http://www2.stat.duke.edu/~rcs46/lectures_2015/01-markdown-git/slides/naming-slides/naming-slides.pdf) consistently (files, objects, columns) - make project self-contained - file organisation - DRY: do not repeat yourself; reuse code by creating functions - documenting code + files (comments, README.md) - consistent coding style See [this post too](http://www2.stat.duke.edu/~rcs46/lectures_2015/01-markdown-git/slides/naming-slides/naming-slides.pdf) and my [personal set of practices I try to follow](https://petrbouchal.xyz/rcode/). ] .pull-right[ ## Tools - version control: git + Github - workflow management: {targets} - testing and validation - software reproducibility: {renv} ] --- class: large, middle, inverse # What next? --- class: large See [workshop page](https://petrbouchal.xyz/czecheval2021) for further resources RStudio [learning resources](https://education.rstudio.com/learn/beginner/) [R for Data Science online book](https://r4ds.had.co.nz/) [\#rstats twitter](https://mobile.twitter.com/search?q=%23rstats&src=hashtag_click) Do an online course Learn git + Github (see [happygitwithr.com/](https://happygitwithr.com/)) Working with Czech public data? Check out packages: {[CzechData](https://jancaha.github.io/CzechData)}, {[RCzechia](https://cran.r-project.org/package=RCzechia)}, {[czso](https://cran.r-project.org/package=czso)} ??? https://docs.google.com/forms/d/e/1FAIpQLSemjKMGKcsML5icIT7wfQ01rP5DiXJsYbr0dwJo3nwRHnIhfw/viewform?usp=sf_link --- class: center, middle, large, inverse [petrbouchal.xyz/czecheval2021](https://petrbouchal.xyz/czecheval2021) --- class: inverse, bottom, right, large layout: false <a href="https://twitter.com/petrbouchal"><svg style="height:0.8em;top:.04em;position:relative;fill:white;" viewBox="0 0 512 512"><path d="M459.37 151.716c.325 4.548.325 9.097.325 13.645 0 138.72-105.583 298.558-298.558 298.558-59.452 0-114.68-17.219-161.137-47.106 8.447.974 16.568 1.299 25.34 1.299 49.055 0 94.213-16.568 130.274-44.832-46.132-.975-84.792-31.188-98.112-72.772 6.498.974 12.995 1.624 19.818 1.624 9.421 0 18.843-1.3 27.614-3.573-48.081-9.747-84.143-51.98-84.143-102.985v-1.299c13.969 7.797 30.214 12.67 47.431 13.319-28.264-18.843-46.781-51.005-46.781-87.391 0-19.492 5.197-37.36 14.294-52.954 51.655 63.675 129.3 105.258 216.365 109.807-1.624-7.797-2.599-15.918-2.599-24.04 0-57.828 46.782-104.934 104.934-104.934 30.213 0 57.502 12.67 76.67 33.137 23.715-4.548 46.456-13.32 66.599-25.34-7.798 24.366-24.366 44.833-46.132 57.827 21.117-2.273 41.584-8.122 60.426-16.243-14.292 20.791-32.161 39.308-52.628 54.253z"/></svg></a> <a href="https://github.com/petrbouchal"><svg style="height:0.8em;top:.04em;position:relative;fill:white;" viewBox="0 0 496 512"><path d="M165.9 397.4c0 2-2.3 3.6-5.2 3.6-3.3.3-5.6-1.3-5.6-3.6 0-2 2.3-3.6 5.2-3.6 3-.3 5.6 1.3 5.6 3.6zm-31.1-4.5c-.7 2 1.3 4.3 4.3 4.9 2.6 1 5.6 0 6.2-2s-1.3-4.3-4.3-5.2c-2.6-.7-5.5.3-6.2 2.3zm44.2-1.7c-2.9.7-4.9 2.6-4.6 4.9.3 2 2.9 3.3 5.9 2.6 2.9-.7 4.9-2.6 4.6-4.6-.3-1.9-3-3.2-5.9-2.9zM244.8 8C106.1 8 0 113.3 0 252c0 110.9 69.8 205.8 169.5 239.2 12.8 2.3 17.3-5.6 17.3-12.1 0-6.2-.3-40.4-.3-61.4 0 0-70 15-84.7-29.8 0 0-11.4-29.1-27.8-36.6 0 0-22.9-15.7 1.6-15.4 0 0 24.9 2 38.6 25.8 21.9 38.6 58.6 27.5 72.9 20.9 2.3-16 8.8-27.1 16-33.7-55.9-6.2-112.3-14.3-112.3-110.5 0-27.5 7.6-41.3 23.6-58.9-2.6-6.5-11.1-33.3 2.6-67.9 20.9-6.5 69 27 69 27 20-5.6 41.5-8.5 62.8-8.5s42.8 2.9 62.8 8.5c0 0 48.1-33.6 69-27 13.7 34.7 5.2 61.4 2.6 67.9 16 17.7 25.8 31.5 25.8 58.9 0 96.5-58.9 104.2-114.8 110.5 9.2 7.9 17 22.9 17 46.4 0 33.7-.3 75.4-.3 83.6 0 6.5 4.6 14.4 17.3 12.1C428.2 457.8 496 362.9 496 252 496 113.3 383.5 8 244.8 8zM97.2 352.9c-1.3 1-1 3.3.7 5.2 1.6 1.6 3.9 2.3 5.2 1 1.3-1 1-3.3-.7-5.2-1.6-1.6-3.9-2.3-5.2-1zm-10.8-8.1c-.7 1.3.3 2.9 2.3 3.9 1.6 1 3.6.7 4.3-.7.7-1.3-.3-2.9-2.3-3.9-2-.6-3.6-.3-4.3.7zm32.4 35.6c-1.6 1.3-1 4.3 1.3 6.2 2.3 2.3 5.2 2.6 6.5 1 1.3-1.3.7-4.3-1.3-6.2-2.2-2.3-5.2-2.6-6.5-1zm-11.4-14.7c-1.6 1-1.6 3.6 0 5.9 1.6 2.3 4.3 3.3 5.6 2.3 1.6-1.3 1.6-3.9 0-6.2-1.4-2.3-4-3.3-5.6-2z"/></svg></a> <a href="https://linkedin.com/in/petrbouchal"><svg style="height:0.8em;top:.04em;position:relative;fill:white;" viewBox="0 0 448 512"><path d="M416 32H31.9C14.3 32 0 46.5 0 64.3v383.4C0 465.5 14.3 480 31.9 480H416c17.6 0 32-14.5 32-32.3V64.3c0-17.8-14.4-32.3-32-32.3zM135.4 416H69V202.2h66.5V416zm-33.2-243c-21.3 0-38.5-17.3-38.5-38.5S80.9 96 102.2 96c21.2 0 38.5 17.3 38.5 38.5 0 21.3-17.2 38.5-38.5 38.5zm282.1 243h-66.4V312c0-24.8-.5-56.7-34.5-56.7-34.6 0-39.9 27-39.9 54.9V416h-66.4V202.2h63.7v29.2h.9c8.9-16.8 30.6-34.5 62.9-34.5 67.2 0 79.7 44.3 79.7 101.9V416z"/></svg></a> petrbouchal [petrbouchal.xyz](https://petrbouchal.xyz) pbouchal@gmail.com